HTG Transcriptome Panel
The HTG Transcriptome Panel (HTP) is expertly designed to provide extensive coverage of most human mRNA transcripts including isoforms. The panel can simultaneously interrogate 19,398 targets using neoplastic and normal Formalin Fixed Paraffin Embedded (FFPE), extracted RNA from FFPE, FF tisue samples and cells, blood samples collected in PAXgene RNA tubes, or untreated cell lines and cell pellets generating data for up to 96 samples in less than three days. HTP utilizes our proprietary workflow and leverages the sensitivity and dynamic range of next-generation sequencing (NGS), allowing researchers to generate reliable results using limited sample amount.
For Research Use Only. Not for Use in Diagnostic Procedures.
Features and Benefits
-
Expertly Curated Panel: Fast and easy to use solution to measure mRNA transcriptome
-
Simplify your Transcriptome Analysis: Generate comprehensive mRNA profile with our streamlined workflow
-
Extraction Free Workflow: Transcriptome analysis from limited amount and low quality RNA samples
-
Compatible with common, biologically relevant sample types including FFPE, PAXgene, extracted RNA and cells
-
Simplified Data Analysis: Provides one click solution to visualization data and generate publication ready figures
Sample Requirements
| Sample Type | Recommended Sample Input |
|---|---|
Larger sample input amounts must be diluted. Please consult the assay package insert and HTG EdgeSeq System User Manual for more details. |
|
| Neoplastic FFPE | 11 mm^2 |
| Normal or any other non-neoplastic FFPE | 22 mm^2 |
| Extracted RNA | 70 ng |
| PAXgene | 400 µL |
| Cells | ≥3,000 cells |
Performance
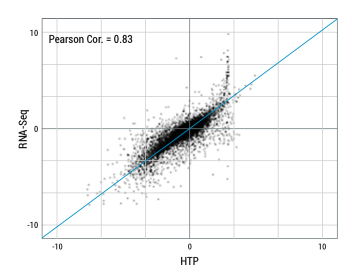
Differential gene expression analysis comparison between HTG Transcriptome Panel and RNA-Seq. HTP shows equivalent results using a fraction of the sample input and is able to generate higher sample pass rate when compared to RNA-Seq
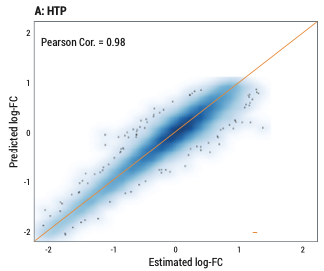
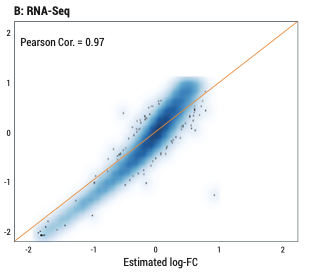
Accuracy of differential expression analysis of HTG Transcriptome Panel and RNA-Seq. Pearson Cor. between the predicted and observed log fold-changes (log-FC) of HTP (A) and RNA-Seq (B).
Precision of HTP Transcriptome Panel
The Precision of HTP was evaluated using samples across multiple plates, operators, processors and lots. Lin's concordance correlation values were high across all conditions demonstrating high precision.
|
Sample |
Mean Lc |
Median Lc |
|---|---|---|
|
Neoplastic FFPE |
0.944 |
0.953 |
|
Normal FFPE |
0.937 |
0.952 |
|
eRNA from FFPE |
0.903 |
0.910 |
|
eRNA from FF |
0.909 |
0.927 |
|
eRNA from Cells |
0.940 |
0.950 |
|
PAXgene |
0.896 |
0.908 |
|
Cells |
0.970 |
0.980 |
Performance of Archival Samples
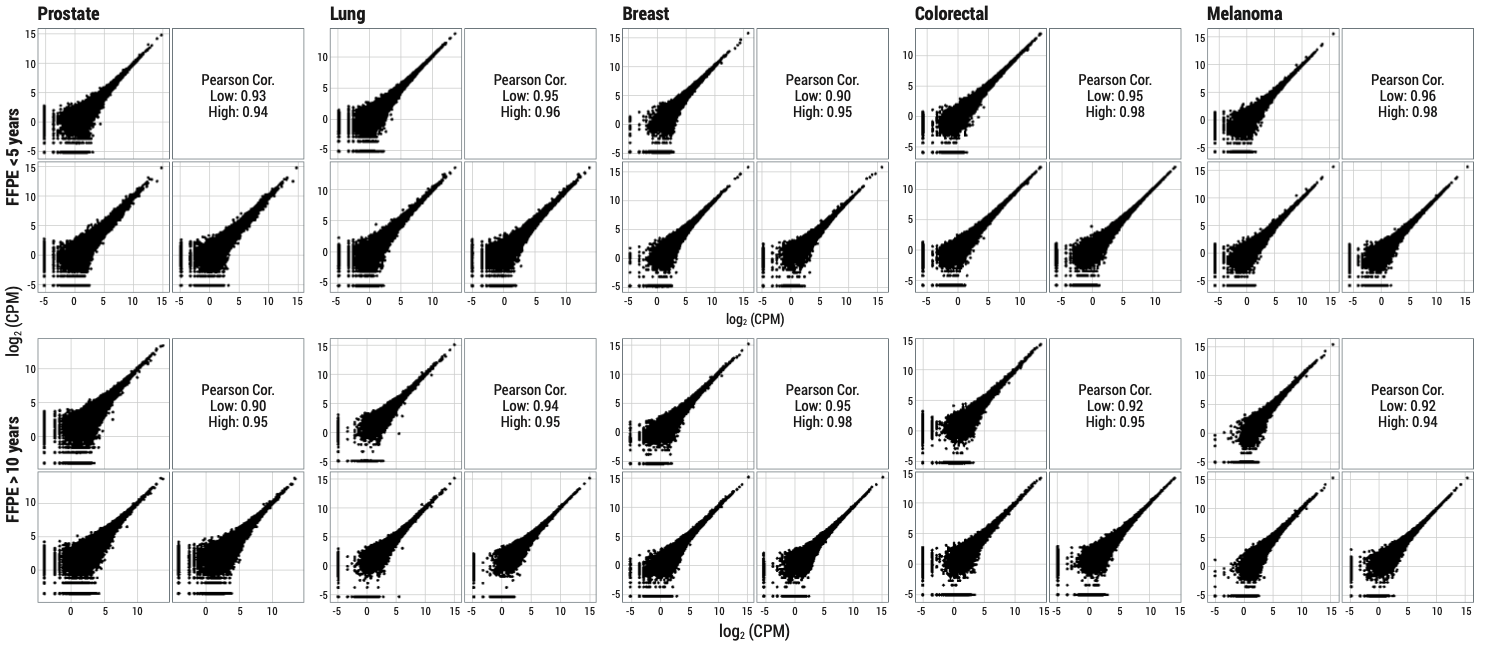
Multi-tissue FFPE samples were lysed and processed with the HTG Transcriptome Panel from blocks less than 5 years old (top row) and blocks greater than 10 years old (bottom row). HTP demonstrates excellent repeatability and offers advantages to RNA-Seq when analyzing archival samples.
Integration with HTG EdgeSeq Reveal software
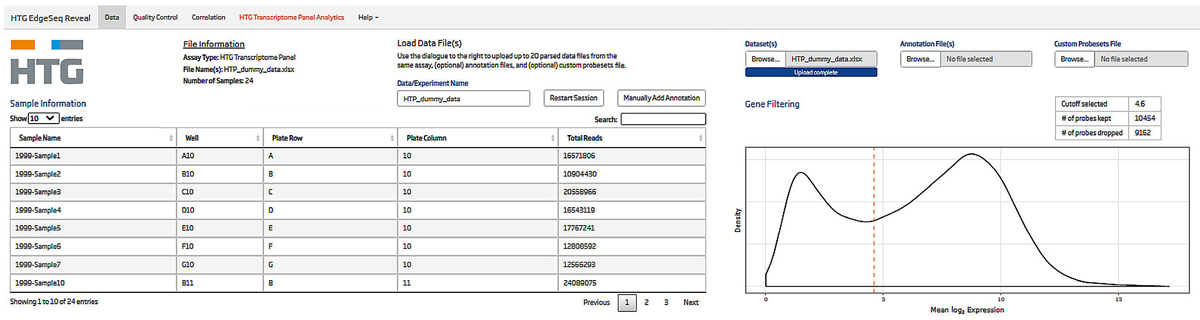
HTG EdgeSeq Reveal software is a powerful, simple-to-use platform for interrogating and visualizing HTG data. The HTG Transcriptome Panel is fully compatible with the HTG EdgeSeq Reveal platform and allows users to gain insights into complex biology by applying QC metrics and using data analysis that enable researchers to gain actionable insights quickly and reliably. Specifically with HTP, there is a user adjustable gene filtering function which allows researchers to exclude genes with low or no expression from further analysis. HTG EdgeSeq Reveal contains an array of valuable biostatistical tools, such as volcano plots, heatmaps and differential gene expression analysis, that can be used to quickly generate meaningful data and reports without a complicated analysis pipeline.
Ordering Information
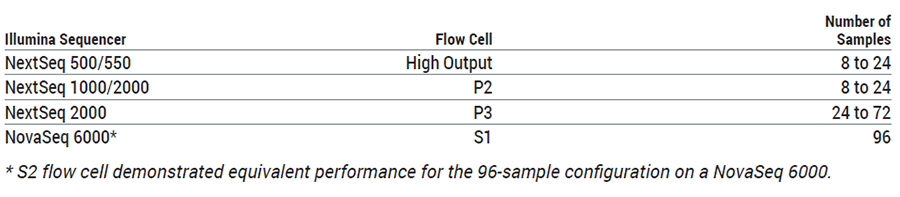
When placing an order, please specify the catalog number. The HTG Transcriptome Panel is compatible with Illumina sequencers
Kit Configurations for use with Illumina Sequencers
HTG-001-008 HTG Transcriptome Panel (4x8)
HTG-001-208 HTG Transcriptome Panel (2x8)
HTG-001-024 HTG Transcriptome Panel (4x24)
HTG-001-224 HTG Transcriptome Panel (1x24)
HTG-001-096 HTG Transcriptome Panel (1x96)
Sequencer Compatibility and Configuration
For Research Use Only. Not for Use in Diagnostic Procedures.
Resources
Learn more about the HTG Transcriptome Panel
Tech Note: HTG Transcriptome Panel Gene Expression Profiling Data Demonstrate High Correlation Using Illumina and Thermo Fisher Sequencing Platforms
Download pdf 223KB
HTG Transcriptome Panel Probe List
Download pdf 378KB
HTG Transcriptome Panel Product Sheet
Download pdf 754KB
Tech Note: Application of HTG Transcriptome Panel for Targeted Gene Expression Profiling in COVID-19 Cases
Download pdf 213KB
Tech Note: Benchmark Study of HTG Transcriptome Panel and NanoString nCounter PanCancer Immune Profiling and Custom Panels
Download pdf 312KB
Publications
View External Link Proteogenomic profiling of high-grade B-cell lymphoma with 11q aberrations and Burkitt lymphoma reveals lymphoid enhancer binding factor 1 as a novel biomarker
View External Link Overexpression of KMT9α Is Associated with Aggressive Basal-like Muscle-Invasive Bladder Cancer
View External Link Blastoid B-Cell Neoplasms: Diagnostic Challenges and Solutions
View External Link EBMT 2022: Acquired Aplastic Anemia Shows Increased Nestin+ Niche Numbers, Upregulation of Genes of the Adaptive Immune System Cells and of CXCL12 in Transformation
Download pdf 1.4MB
ASH 2022: 4144: Bone Marrow Fibrosis Is Associated with Non-Response to CD19 CAR-T Therapy
Download pdf 2.0MB
View External Link ASH 2022: 2859: Transcriptomic Comparison of Non-Hodgkin Lymphomas in Relapsed/Refractory Versus Newly Diagnosed Patients Using Single FFPE Slides
Download pdf 697KB
View External Link Transcriptome-Wide Gene Expression Profiles from FFPE Materials Based on a Nuclease Protection Assay Reveals Significantly Different Patterns between Synovial Sarcomas and Morphologic Mimickers
View External Link Mesenchymal-epithelial transition in lymph node metastases of oral squamous cell carcinoma is accompanied by ZEB1 expression
View External Link DLBCL Cell of Origin Typing and Whole Transcriptome Analysis using Single Slides with HTG EdgeSeq
Download pdf 1.8MB
CK5/6 and GATA3 Defined Phenotypes of Muscle-Invasive Bladder Cancer: Impact in Adjuvant Chemotherapy and Molecular Subtyping of Negative Cases
Download pdf 4.8MB
View External Link Avelumab maintenance in advanced urothelial carcinoma: biomarker analysis of the phase 3 JAVELIN Bladder 100 trial
View External Link
White Papers
Gene Expression Profiling and Sample Type Comparison
between the HTG Transcriptome Panel and RNA-Seq
Download pdf 415KB
Overview of the Design and Performance of the HTG Transcriptome Panel
Download pdf 1.5MB
Comparison of Prototype HTG Transcriptome Panel
Download pdf 928KB
Proof-of-Concept for a Whole Transcriptome Panel
Using HTG EdgeSeq™ Technology
Download pdf 192KB
Learn More
For further information on the HTG Transcriptome Panel, please fill out the registration form below. Or call us at (877) 507-3259.
Page last updated January 04, 2023